Output files
Here, using the results produced in the Non-bacterial dataset section, we give users a glimpse over the main outputs produced by ngs-preprocess. The command used in the quickstart wrote the results under the preprocessed_reads directory.
Note
Please take note that the pipeline uses the directory set with the --output parameter as a storage place in which it will create a folder for the final pre-processed reads and for the intermediate files, separated by sequencing technology.
Directory tree
After a successful execution, you will have something like this:
# Directory tree from the running dir
preprocessed_reads
# directory containing the final results of the data cleaning
├── final_output
│ └── nanopore
│ └── SRR23337893.filtered.fq.gz
# a template input ready for MpGAP
├── mpgap_samplesheet.yml
# directory containing the nextflow execution reports
├── pipeline_info
│ ├── ngs_preprocess_report_2023-11-18_10-07-36.html
│ ├── ngs_preprocess_timeline_2023-11-18_10-07-36.html
│ ├── ngs_preprocess_tracing_2023-11-18_10-07-36.txt
# directory containing the intermediate files produced by the tools used during pre-processing, and, QC
├── preprocessing_outputs
│ └── nanopore
│ ├── porechop
│ └── QC
# directory containing the intermediate files when downloading data from SRA
└── SRA_FETCH
├── FASTQ
│ └── SRR23337893_data
└── SRR23337893_sra_runInfo.csv
The pre-formatted MpGAP input samplesheet
Once finished, the pipeline also generates a file called mpgap_samplesheet.yml (showed below). Basically this samplesheet defines all the minimum definitions in order to assemble these reads using the MpGAP pipeline.
samplesheet:
- id: SRR23337893
nanopore: /workspace/ngs-preprocess/testing/preprocessed_reads/final_output/nanopore/SRR23337893.filtered.fq.gz
Note
One must keep in mind that, this template samplesheet contains only the bare minimum to launch MpGAP but many other customizations are possible. For example, the generated samplesheet will assemble each read separately, but, MpGAP can also perform hybrid assemblies. Therefore, users can/must use this output as a template for easily customization of the assembly pipeline input to use the results of ngs-preprocess pipeline.
For more information, please refer to the MpGAP documentation.
Example of QC outputs
Here I am going to display just a very few examples of results produced, focusing on the QC, as the main result is a cleaned FASTQ file.
Length versus Quality Scatterplot
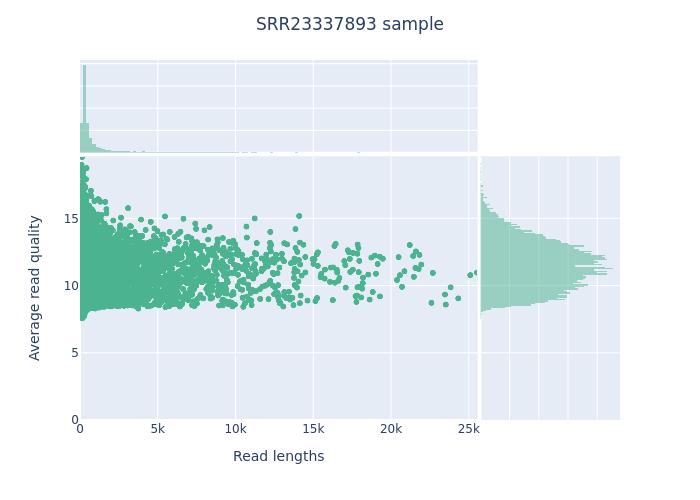
NanoPlot Report HTML
Open it here.
NanoStats Report TXT
Open it here.